Cracking the sugar code: Why the 'glycome' is the next big thing in health and medicine
When you think of sugar, you probably think of the sweet, white, crystalline table sugar that you use to make cookies or sweeten your coffee. But did you know that within our body, simple sugar molecules can be connected together to create powerful structures that have recently been found to be linked to health problems, including cancer, aging and autoimmune diseases.
These long sugar chains that cover each of our cells are called glycans, and according to the National Academy of Sciences, creating a map of their location and structure will usher us into a new era of modern medicine. This is because the human glycome – the entire collection of sugars within our body – houses yet-to-be-discovered glycans with the potential to aid physicians in diagnosing and treating their patients.
Thanks to the worldwide attention garnered by the 2003 completion of the Human Genome Project, most people have heard about DNA, genomics and even proteomics – the study of proteins. But the study of glycans, also known as glycomics, is about 20 years behind that of other fields. One reason for this lag is that scientists have not developed the tools to rapidly identify glycan structures and their attachment sites on people’s cells. The “sugar coat” has been somewhat of a mystery.
Until now, that is.
While most laboratories focus on cellular or molecular research, our lab is dedicated to developing technology to rapidly characterize glycan structures and their attachment sites. Our ultimate goal is to catalog the hundreds of thousands of sugars and their locations on various cell types, and then to use this information to tailor medical therapies to each individual.
Why do we care about glycans?
In the future, it is likely that analysis of an individual’s glycans will be used to predict our risk for developing diseases like rheumatoid arthritis, cancer or even food allergies. This is because glycome alterations can be specifically tied to particular disease states. Also, biological processes like aging are linked to inflammation in our glycome. It remains to be tested if reversing these changes can help prevent disease, or even slow aging – an intriguing possibility.
Along with DNA, proteins, and fats, glycans are one of the four major macromolecules essential for life. Of these four, glycans are the final arbiters of how our cells behave.
DNA orchestrates what we look like, our capacity to think and behave, and even determines the diseases to which we are most susceptible. Within our DNA are short segments, genes, which often contain instructions for how to synthesize proteins. Proteins in turn are the “workhorses” of the cell, carrying out many of the functions necessary for life.
However, how a protein behaves often depends on what glycans are attached to it. In other words, these sugar molecules can greatly influence how our proteins do their work, and even how our cells will respond to stimuli. For example, if you change a few glycans on the outside of a cell, it might trigger that cell to migrate to a different location in our body.
The main job of glycans is to modify the proteins and fats that sit on the surface of our cells. Together, they create a thick sugar coat around the cell. If we consider the surface of the cell to be soil, then glycans would be the wonderfully diverse plant-life and foliage that sprout up and bring color and identity to the cell. In fact, if you were able to see a cell with your naked eye, it would look very fuzzy. Picture a peach with 10 times more fuzz.
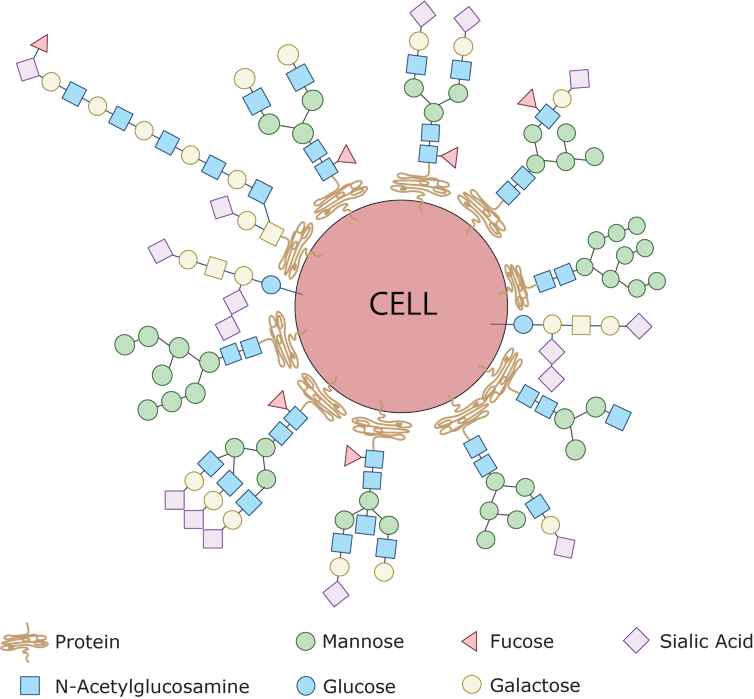
Glycans label our own cells and identify them as ‘self’
The fuzz around a cell is its glycan coat. Being on the outside of our cells, glycans are the first point of contact for most cellular interactions and thus influence how our cells communicate with one another. You can also think of the glycans as a unique cellular “barcode.” Thus, a kidney cell’s fuzz will look different from an immune cell’s fuzz. But there are also similarities. In fact, the immune cells that survey our body searching for pathogens know not to attack our own “self” cells because of common features in the glycan “barcode” which are shared by all cells of our body.
In contrast, bacteria and parasites like malaria have different “sugar coats” that are not seen on human cells. When bacterial sugars are tagged as “foreign,” a person’s immune system targets the bacterium for destruction. However, some harmful bacterial pathogens like group B streptococcus, which commonly cause severe infections in babies, can avoid immune detection by impersonating human cells by carrying similar glycans as a disguise – like the wolf dressed in sheepskin.
Unfortunately some pathogens are also able to use our glycans to help them cause disease. Deadly viruses like HIV and Ebola have evolved to grab hold of specific glycans which they then “lock” onto as they infect our human cells. Therapies that either block these viruses from interacting with our glycans, or that attack virus-specific glycans may be a new avenue to treating these infections.
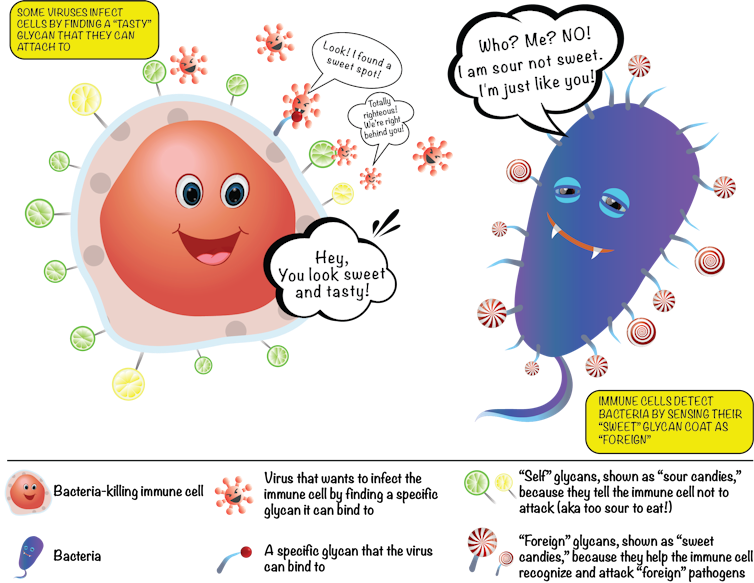
New research has also shown that glycans play a huge role in the development of autoimmune diseases like rheumatoid arthritis and autoimmune pancreatitis. This is not surprising since glycans directly influence the function of immune cells.
Normally, our immune cells act as our body’s “defense system,” and identify and destroy foreign invaders like harmful bacteria or viruses. But when the body mistakenly labels our own cells as the enemy and launches an internal attack on itself, autoimmunity is born. Interestingly, in such instances, it is the glycans present on the misbehaving self-attacking antibodies that will dictate the strength of the attack on the body. This abnormal immune response can even be directed against glycans. For example, the immune system can mistake “self” glycans as if they were “foreign” molecules. Our research team recently published an article that introduced the glycan theory of autoimmunity, which explains some of these relationships.
Glycans in our food can trigger immune responses
There have been many studies linking consumption of red meat with diseases like atherosclerosis and diabetes, but they have not been able to show why or how this occurs until recently. One intriguing study suggests that the culprit was a sugar with the unwieldy name, nonhuman sialic N-glycolylneuraminic acid, or Neu5Gc for short. Neu5Gc is found in all mammals except humans, because the early humans that could make Neu5Gc died from an ancient malarial parasite.
However, although we now lack the ability to produce Neu5Gc, our bodies still have the ability to incorporate it into the glycans on our cells if we obtain it by eating red meat. Once it becomes part of our cells’ glycan coat, our cells then have a “foreign” substance – Neu5Gc – surrounding them. This can trigger inflammation throughout the body because our immune system recognize Neu5Gc as “foreign” and attacks it. The chronic inflammation caused by these internal attacks can lead to heart attack, stroke and even cancer.
Our bodies synthesize tens of thousands of unique glycans, often with branching structures formed from simple sugar building blocks. Proteins or fats can also be modified by dozens of unique glycans. These countless combinations make mapping glycans a difficult task because we need a practical and efficient way to analyze hundreds of thousands of glycan patterns.
Our research team has now developed methods to rapidly and robustly monitor the human glycome. By capitalizing on engineering advancements and improvements in sample processing, our technique can monitor thousands of glycans at once, which allows us to characterize the glycans in cells from healthy controls and patients with a variety of different diseases. Our goal is to use this data to develop predictive models to help clinicians diagnose and treat all human diseases. We believe that a new wave of medical advancements will arrive as we unlock the “sugar code.”
Jenny Wang was the co-lead author of this article.![]()
Emanual Maverakis, Associate Professor- Departments of Medical Microbiology & Immunology and Dermatology | Member- Foods For Health Institute | Member- Comprehensive Cancer Center | Director- Autoimmunity | Director- Immune Monitoring Core, University of California, Davis; Carlito Lebrilla, Distinguished Professor of Chemistry, University of California, Davis, and Jenny Wang, Clinical Research Fellow, University of California, Davis | Medical Student, Albert Einstein College of Medicine, Yeshiva University
This article was originally published on The Conversation. Read the original article.