Uncovering insights hidden within the psoriasis transcriptome.
Herein we explain how our novel data mining strategies for RNA-Seq datasets lead to new insights into the pathophysiology of psoraisis, an immune-mediated disease involving the skin. This story can also be found in our recently published manuscript in JCI Insight. You can click here to read the entire publication (subscription NOT required).Introduction
The skin transcriptome is the sum of all messenger RNA molecules expressed in the skin. About a decade ago, next generation DNA sequencing technologies allowed researchers to investigate the skin transcriptome in the setting of psoriasis. This led to the discovery of thousands of genes that were differentially expressed in psoriasis skin; But are all of these genes involved in psoriasis pathophysiology? As is the case with other diseases, knowing the sum of all expressed genes in psoriasis is only a small piece of the puzzle. How to decipher the psoriasis transcriptome has been a challenge that our research team and others are still working on. One strategy to narrow down the thousands of differentially genes to a more reasonable number is to set a threshold as to what represents a meaningful up or down regulation in gene expression. For example, is it meaningful if a gene's expression is increased by only 1.2 fold in the setting of psoriasis?
Meaningfulness is different from significance, which simply means that the observed change is not likely to be due chance. When analyzing DNA sequencing data for significance, p values need to be adjusted for multiple testing, as transcriptome datasets have thousands of genes. Programs used for such tasks take many factors into consideration (number of uniquely mapping reads, total number of genes monitored, variation in read counts etc). However, even when thresholds for significance and meaningfulness are met, investigators can easily be led astray - as data crunching by these classical methods often over or under emphasize the importance of a gene.
The error of meaningfulness thresholds
It is common to set a 2-fold increase or decrease in gene expression as a threshold for meaningfulness. However, is this the best way to identify differentially expressed genes of interest? It depends on the gene, its expression level, and the situation. Lets take for example TNF. During the 1990's, an area of immunology when virtually all cytokines were categorized as belonging to either Th1 or Th2 immune responses, TNF and IFN-g were viewed as the classical Th1 cytokines. This led to the testing of anti-IFN-g and anti-TNF-based therapies in animal models of autoimmunity. In such experiments, anti-TNF therapies showed superior efficacy. In contrast, anti-IFN-g treatments sometimes made autoimmunity paradoxically more severe. Eventually, anti-TNF therapies also proved useful in treating a variety of human autoimmune diseases. With regards to psoriasis, anti-TNF biologic therapies are indeed effective treatments. Although newer psoriasis biologic treatments target different molecules, the prototypic anti-TNF agent, infliximab, is still arguable one of the fastest and most effective medications for psoriasis (in terms of skin clearing at 10 weeks). However, the effectiveness of anti-TNF-based therapies would not have been predicted from TNF gene expression in psoriasis plaques. In comparison to healthy control skin or uninvolved skin, TNF expression within a psoriatic plaque is only marginally increased. In reality, TNF is made by numerous cell types not simply Th1 cells and relatively small changes in its expression have dramatic effects on tissue homeostasis. Thus, TNF blockade has dramatic effects on pathogenic immune responses. Setting a 2-fold change as a threshold for meaningfulness would absolutely miss important genes such as TNF.
For the next example lets consider a hypothetical gene expressed by T cells. If in the setting of psoriasis the expression of this hypothetical gene drops by 30% in T cells, its expression in psoriatic skin will paradoxical not be decreased at all. In fact, its tissue expression will rise more than 2-fold, which may lead some investigators to erroneously conclude that the "up-regulated" gene is involved in the pathophysiology of psoriasis. In this example, the hypothetical gene is decreased in T cells but increased in psoriasis plaques. The increased expression is a result of the simple fact that psoriatic plaques have more T cells not that the expression of the gene is otherwise increased. It is likely that the vast majority of the upregulated genes in psoriasis are a mere consequence of the infiltrating immune cells or proliferation of existing cells within the skin. These are just some examples of why deciphering transcriptomic data is difficult. One method we have chosen to overcome some of these issues is to look at relative gene expression, that is the expression of one gene relative to reference immune genes, especially the genes that encode immune receptors such as the T cell receptor genes. Another strategy is to do single cell sequencing. Both strategies have advantages and disadvantages. For example, the cell preparation techniques used for single cell sequencing will alter the cellular transcriptome.
T cell receptors and T cell repertoire analysis
As just stated, we often use TCR genes as reference genes when characterizing an immune response. However, this strategy has a variety of barriers. TCRs are encoded by productive rearrangements of the alpha beta or gamma delta TCR genes. For the beta and delta chains, the genetic recombination events involve variable (V), diversity (D), joining (J), and constant (C) region exons. This allows for the immune system to express an almost infinite number of immune receptors to project of from invading pathogens and cancer.
Traditionally, TCR genes are sequenced directly from the products of TCR-specific PCR reactions. TCR mining of RNA-Seq datasets is an alternative approach to characterize T cell repertoires, but requires the use of an appropriate bioinformatics alignment pipeline. In our recent JCI Insight manuscript we developed and validated a pipeline to estimate the overall and relative expression of TCR V/J/C segments. Our method has the advantage over traditional TCR repertoire analysis in that it allows investigators to conduct correlative studies between TCR genes and other genes of interest; this is because it detects all TCR-mapping reads in an RNA-Seq dataset. In contrast, bioinformatics tools focused solely on analysis of the TCR CDR3 region cannot accurately estimate TCR gene segment usage and CDR3-mapping only yields a small handful of reads per RNA-Seq dataset, making correlative studies difficult. Also, traditional RNA-Seq analysis pipelines are not aware of TCR rearrangements and may not take into account gene rearrangements, considering them low quality mapping reads.
Our approach allowed us to not only identify novel psoriasis-associated TCR gene segments, but also correlate the transcription of individual alpha beta and gamma delta TCR gene segments with genes of known importance to the pathophysiology of psoriasis. Four independently acquired RNA-Seq sample sets allowed for rigorous statistical validation of our results.
Our manuscript highlights a many exciting findings. For example, by focusing on the expression of individual TCR gene segments relative to all TCR gene segments of the same family, we discovered that TRAJ23 is proportionally over expressed and TRAJ39 is proportionally under expressed in psoriasis (Bonferroni p = 6.19 x 10e-27 and 1.43 x 10e-248, respectively). (TRAJ23 and TRAJ39 are TCR alpha gene segments). We also found that a TCR gamma gene segment is minimally over expressed in psoriasis, TRGV5 (Bonferroni p = 6.03 x 10e-13)). A finding that we were able to validate across 4 different RNA-Seq datasets. The figure below summarizes these results. Please see the full manuscript for more details. This same strategy can be applied to other autoimmune and cancer datasets, and thus carries wide applications. Importantly, our results differ from the alpha beta gene segments previously reported to be disproportionately overrepresented in psoriasis (1-3). These highly conflicting results may be due to the exceedingly small sample sizes used in the prior studies, which for the most part lacked summary statistics to evaluate the significance of the psoriasis-associated TCR genes.
T cells, IL-17A, and psoriasis
Since the demonstrated efficacy of cyclosporine in psoriasis (4), alpha beta T cells have been thought to be central to psoriasis pathophysiology, and recent clinical evidence points to the Th17 subtype as the driver of disease (5), as is evident by the success of IL-17/IL-23-targeted therapies (6-9). However, many groups are pursuing other sources of IL-17 as important mediators of the psoriatic phenotype. For example, some investigators have posited that in psoriasis, gamma delta T cells are expanded disproportionately more than alpha beta T cells, and that the former is the dominant source of IL-17 (10-12).
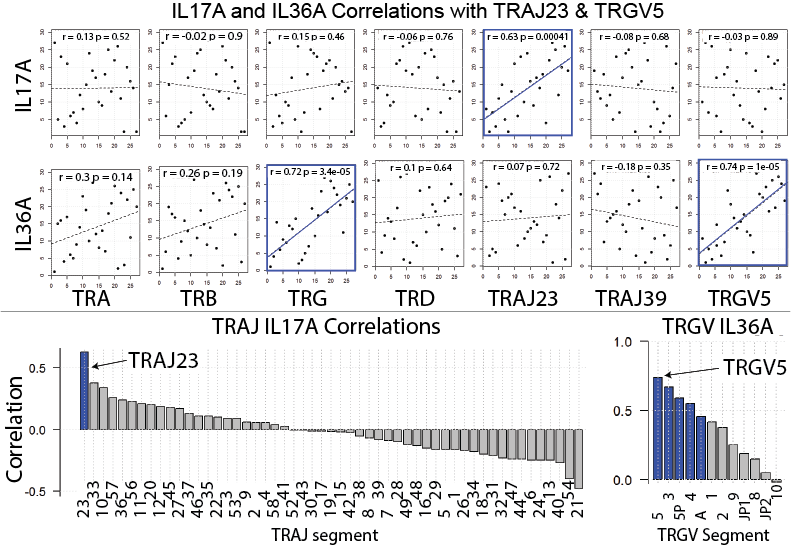
To address the debate of alpha beta versus gamma delta T cells head on we compared the relative frequencies of the corresponding alpha, beta, gamma, delta gene segments in psoriatic and normal skin (surprisingly nobody has done this before). By looking at relative gene expression, we found no evidence for a differential expansion of gamma delta T cells over alpha beta T cells. In fact, gamma delta TCR gene segments were only marginally over expressed in the setting of psoriasis. We next sought to address the source of IL-17 by plotting the expression of individual TCR gene segments against IL17A expression. Again, we found no evidence that gamma delta TCR gene segments correlated with IL17A expression. Interestingly, TRGV5 expression did correlate with IL36. In contrast, TRAJ23 expression correlated well with IL17A. These results were then validated in four independently collected RNA-Seq datasets. From these results we speculate that TRAJ23-expressing cells (abundant in psoriasis) are a source of IL-17 and TRGV5-expressing T cells possibly expand in response to IL-36, a psoriasis-associated cytokine. These results are summarized in the figure below. For more details of this study and to see our other exciting discoveries please see the published manuscript by clicking here.
1. Chang YT, Liu HN, Shiao YM, Lin MW, Lee DD, Liu MT, et al. A study of PSORS1C1 gene polymorphisms in Chinese patients with psoriasis. The British journal of dermatology. 2005;153(1):90-6.
2. Menssen A, Trommler P, Vollmer S, Schendel D, Albert E, Gurtler L, et al. Evidence for an antigen-specific cellular immune response in skin lesions of patients with psoriasis vulgaris. Journal of immunology. 1995;155(8):4078-83.
3. Vollmer S, Menssen A, and Prinz JC. Dominant lesional T cell receptor rearrangements persist in relapsing psoriasis but are absent from nonlesional skin: evidence for a stable antigen-specific pathogenic T cell response in psoriasis vulgaris. The Journal of investigative dermatology. 2001;117(5):1296-301.
4. Ellis CN, Fradin MS, Messana JM, Brown MD, Siegel MT, Hartley AH, et al. Cyclosporine for plaque-type psoriasis. Results of a multidose, double-blind trial. N Engl J Med. 1991;324(5):277-84.
5. Thaci D, Blauvelt A, Reich K, Tsai TF, Vanaclocha F, Kingo K, et al. Secukinumab is superior to ustekinumab in clearing skin of subjects with moderate to severe plaque psoriasis: CLEAR, a randomized controlled trial. J Am Acad Dermatol. 2015;73(3):400-9.
6. Langley RG, Elewski BE, Lebwohl M, Reich K, Griffiths CE, Papp K, et al. Secukinumab in plaque psoriasis--results of two phase 3 trials. N Engl J Med. 2014;371(4):326-38.
7. McInnes IB, Mease PJ, Kirkham B, Kavanaugh A, Ritchlin CT, Rahman P, et al. Secukinumab, a human anti-interleukin-17A monoclonal antibody, in patients with psoriatic arthritis (FUTURE 2): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2015;386(9999):1137-46.
8. Papp KA, Leonardi C, Menter A, Ortonne JP, Krueger JG, Kricorian G, et al. Brodalumab, an anti-interleukin-17-receptor antibody for psoriasis. N Engl J Med. 2012;366(13):1181-9.
9. Gordon KB, Blauvelt A, Papp KA, Langley RG, Luger T, Ohtsuki M, et al. Phase 3 Trials of Ixekizumab in Moderate-to-Severe Plaque Psoriasis. N Engl J Med. 2016;375(4):345-56.
10. Cai Y, Shen X, Ding C, Qi C, Li K, Li X, et al. Pivotal role of dermal IL-17-producing gammadelta T cells in skin inflammation. Immunity. 2011;35(4):596-610.
11. Yoshiki R, Kabashima K, Honda T, Nakamizo S, Sawada Y, Sugita K, et al. IL-23 from Langerhans cells is required for the development of imiquimod-induced psoriasis-like dermatitis by induction of IL-17A-producing gammadelta T cells. J Invest Dermatol. 2014;134(7):1912-21.
12. Hartwig T, Pantelyushin S, Croxford AL, Kulig P, and Becher B. Dermal IL-17-producing gammadelta T cells establish long-lived memory in the skin. Eur J Immunol. 2015;45(11):3022-33.